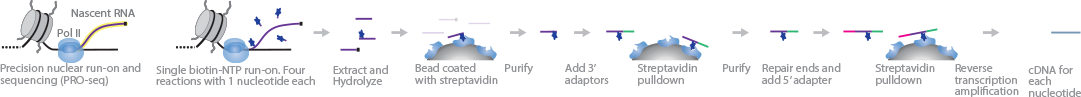New Chromatin Run-On Reaction Enables Global Mapping of Active RNA Polymerase Locations in an Enrichment-free Manner | ACS Chemical Biology

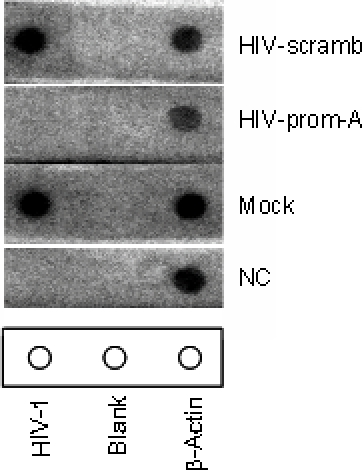
Analysis of Viral Promoter Usage in EBV-Infected Cell Lines: A Comparison of qPCR Following Conventional RNA Isolation and Nuclear Run-On Assay | SpringerLink
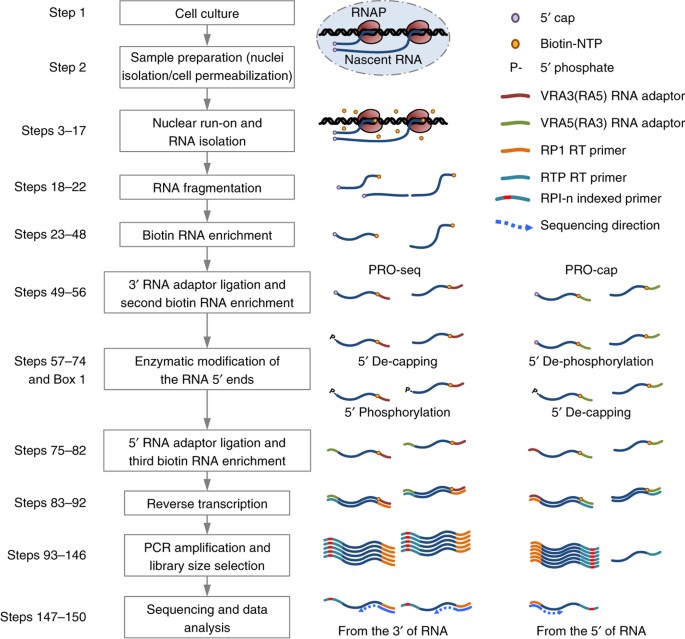
Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq) | Nature Protocols
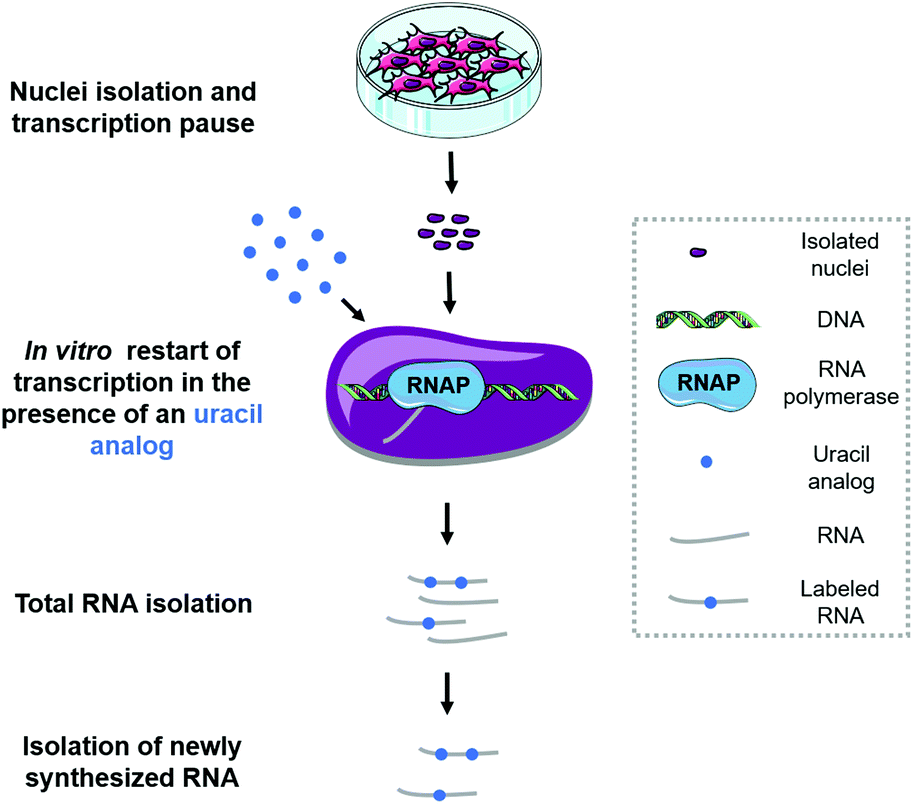
Methods for the analysis of transcriptome dynamics - Toxicology Research (RSC Publishing) DOI:10.1039/C9TX00088G

Quantification of nascent transcription by bromouridine immunocapture nuclear run-on RT-qPCR | Nature Protocols

Standardization of nuclear run-on assay (a) 32P UTP incorporated total... | Download Scientific Diagram
Time-Dependent c-Myc Transactomes Mapped by Array-Based Nuclear Run-On Reveal Transcriptional Modules in Human B Cells | PLOS ONE